Data alignment in time (MJD)
The purpose of this section is to explain how two DataFrame can
be aligned in time.
One may need this functionality in order to apply mathematical operations on two different sets of data both containing timestamps.
What happens in the background
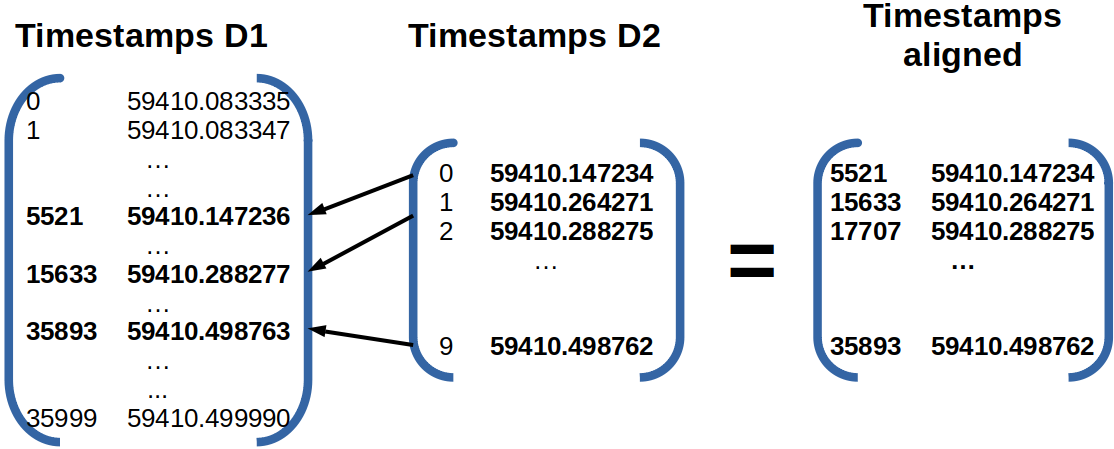
The figure below illustrates how the set D2 is aligned with respect to set D1.
Let’s define two DataFrame D1 and D2, both having an “mjd” column of timestamps in MJD format.
Using the alignDatasets() function, the indexes of D1 will be assigned
to D2 based on their respective timestamps.
In the end, only the alignable rows remain in D2.
Accessing DataFrame
Throughout this part of the tutorial, we will use two White-Rabbit Module
objects. Let’s pick them up.
First, as usual, we create an Infrastructure instance
>>> import palantir.structures as structures
>>> # UTC
>>> startDate = '231102'
>>> endDate = '231102'
>>> startTime = '000000'
>>> endTime = '100000'
>>> infraID = 1
>>> REFIMEVE = structures.Infrastructure(infraID, startDate, endDate, startTime, endTime, 'UTC')
Then, we retrieve our two White-Rabbit Module
objects and get data as DataFrame.
>>> # module1
>>> module1_id = 12
>>> module1 = REFIMEVE.processModule(module1_id)
>>> MOD1 = module1.clock_out.timeChunk.getAsDataframe()
>>> # module2
>>> module2_id = 14
>>> module2 = REFIMEVE.processModule(module2_id)
>>> MOD2 = module2.clock_out.timeChunk.getAsDataframe()
Align two DataFrames
Finally, using the alignDatasets() function, we align
MOD2 with respect to MOD1 timestamps.
>>> import palantir.alignment as alignment
>>> MOD1_aligned, MOD2_aligned = alignment.alignDatasets(MOD1, MOD2, 1, 1, diffMax=0.2)
Possible Troubleshooting
You may encounter a situation where aligning two DataFrame returns empty objects.
One option might be the value of the diffMax parameter in the alignDatasets() function.
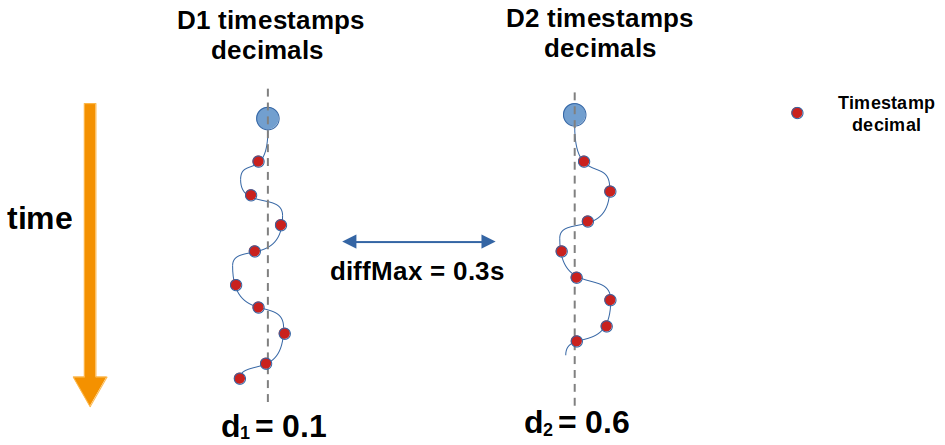
To ilustrate it, see the figure below. We have two datasets D1 and D2, whose timestamps decimal values oscillate respectively around d1=0.1 and d2=0.6.
If diffMax is fixed to 0.3 seconds, no timestamp will fulfill the criteria | mjd2 - mjd1 | < 0.3, leading to empty alignment.